
Whole-genome precision – see the full story of cancer
CancerVision is a tumor-normal paired target-enhanced whole-genome cancer profiling test, providing 99% sensitivity, accurate complex variant detection and accurate genome-wide signatures — all in 1 test.
Target-enhanced WGS
Merging benefits of targeted panel (600+ genes with 500x coverage) plus whole genome sequencing
2-in-1
Whole genome coverage both for somatic (40x) and germline (20x)
>99%
Sensitivity/Positive predictive value*
*>99% for SNVs, >98% for Indels
Accurate complex somatic variants
CNV, SV, non-coding areas
Genome-wide mutational pattern
TMB, MSI, HRD – reported for all tumor types
Custom whole genome analysis
On-demand – e.g., ecDNA, tumor ploidy, transposable elements
CAP/CLIA assay
within a 2 week turnaround time
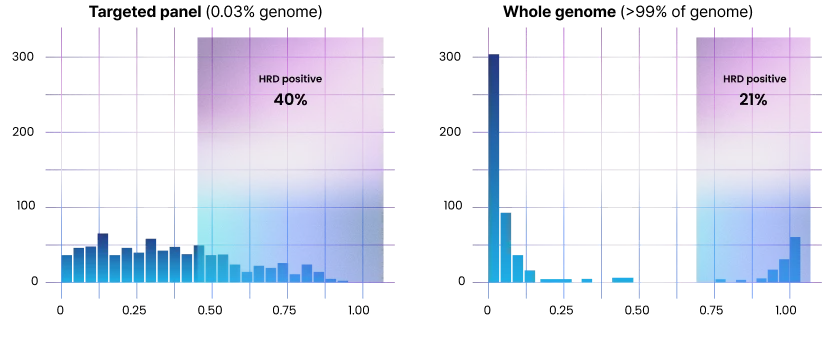
HRD scores based on CancerVision vs. a targeted panel
2 Ju, Y. S et al, (2024). Whole-Genome analyses of 1,364 breast cancers with clinical records. Research Square(Preprint). https://doi.org/10.21203/rs.3.rs-5094752/v1
More accurate genome-wide markers – HRD example from 1,364 breast cancer case
CancerVision’s whole-genome method enables clearer distinction between HRD-positive and HRD-negative, avoiding the ambiguity of targeted panel results.2 Genome-wide marker analysis (including HRD) is not an add-on but a part of CancerVision analysis, at no additional cost.
Accelerate & elevate your research with CancerVision
For cancer researchers
Complete genomic profiling
Identify information that may be potentially missed by WES/TPS
Genome-wide analysis for instability markers
Get a comprehensive view of genomic instability markers with an all-inclusive whole-genome snapshot for accurate detection of TMB, MSI and HRD status
Tumor-specific alterations vs. inherited sequences
Distinguish between tumor-specific vs. inherited genetic alterations across diverse populations germline
Custom analysis for whole-genome biomarkers
Bioinformatics support for whole genome based biomarkers – whole genome doubling, ecDNA, tumor ploidy, MATH score, and more.
For biopharma
Novel biomarker discovery
Leverage whole genome data to uncover previously invisible biomarkers, including genome-wide mutational signatures and non-coding alterations
Novel patient subgroups
Identify responder and non-responder populations for targeted therapies using whole genome insights, enabling more precise patient stratification and higher trial success rates
Accelerated clinical trial enrollment
Rapidly identify and enroll patients for clinical trials by capturing mutations of interest across SNVs, indels, CNVs, SVs, and non-coding regions—speeding up trial timelines
Future-proofed trials
Access comprehensive whole genome data, ensuring readiness for post-phase II/III analyses without the need for additional samples or sequencing
Accepted samples and sample requirements
| Somatic specimen | Volume | Shipping condition |
|---|---|---|
| FFPE tissue | ≥10 from curls or slides in 5 micron thickness, with an H&E stained slide for reference | Ambient |
| Fresh frozen | 25mg of cryopreserved tissue | Frozen or dry ice |
| Germline specimen | Volume | Shipping condition |
|---|---|---|
| Whole blood | 1 Streck or EDTA tube – minimum of 2mL | Ambient or with cold packs |
| gDNA | ≥100ng – minimum concentration of 2ng/uL | Ambient or frozen |
| Saliva | 1 tube in collection device | Ambient |
| Buccal swab | 1 swab in fixative solution | Ambient |